Note
Click here to download the full example code
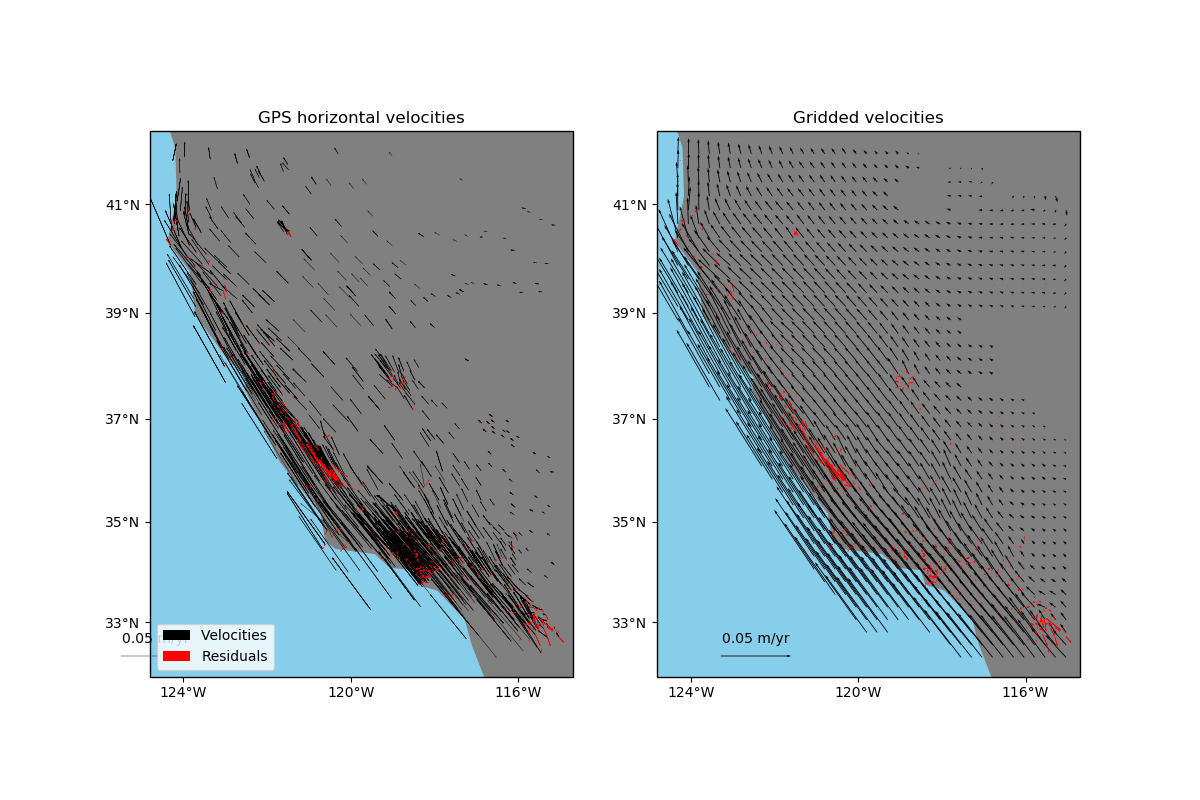
Gridding 2D vectors (coupled)¶
One way of gridding vector data would be grid each component separately using
verde.Spline and verde.Vector. Alternatively,
verde.VectorSpline2D can grid two components simultaneously in a way that
couples them through elastic deformation theory. This is particularly suited, though not
exclusive, to data that represent elastic/semi-elastic deformation, like horizontal GPS
velocities.
Out:
Cross-validation R^2 score: 0.97
/home/travis/miniconda/envs/testing/lib/python3.6/site-packages/cartopy/mpl/geoaxes.py:1752: RuntimeWarning: invalid value encountered in less
u, v = self.projection.transform_vectors(t, x, y, u, v)
/home/travis/miniconda/envs/testing/lib/python3.6/site-packages/cartopy/mpl/geoaxes.py:1752: RuntimeWarning: invalid value encountered in greater
u, v = self.projection.transform_vectors(t, x, y, u, v)
/home/travis/build/fatiando/verde/examples/vectorspline2d.py:128: UserWarning: Tight layout not applied. The left and right margins cannot be made large enough to accommodate all axes decorations.
plt.tight_layout()
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import numpy as np
import pyproj
import verde as vd
# Fetch the GPS data from the U.S. West coast. We'll grid only the horizontal components
# of the velocities
data = vd.datasets.fetch_california_gps()
coordinates = (data.longitude.values, data.latitude.values)
region = vd.get_region(coordinates)
# Use a Mercator projection because VectorSpline2D is a Cartesian gridder
projection = pyproj.Proj(proj="merc", lat_ts=data.latitude.mean())
# Split the data into a training and testing set. We'll fit the gridder on the training
# set and use the testing set to evaluate how well the gridder is performing.
train, test = vd.train_test_split(
projection(*coordinates), (data.velocity_east, data.velocity_north), random_state=0
)
# We'll make a 15 arc-minute grid in the end.
spacing = 15 / 60
# Chain together a blocked mean to avoid aliasing, a polynomial trend to take care of
# the increase toward the coast, and finally the vector gridder using Poisson's ratio
# 0.5 to couple the two horizontal components.
chain = vd.Chain(
[
("mean", vd.BlockReduce(np.mean, spacing * 111e3)),
("trend", vd.Vector([vd.Trend(degree=1) for i in range(2)])),
("spline", vd.VectorSpline2D(poisson=0.5, mindist=10e3)),
]
)
# Fit on the training data
chain.fit(*train)
# And score on the testing data. The best possible score is 1, meaning a perfect
# prediction of the test data.
score = chain.score(*test)
print("Cross-validation R^2 score: {:.2f}".format(score))
# Interpolate our horizontal GPS velocities onto a regular geographic grid and mask the
# data that are far from the observation points
grid_full = chain.grid(
region, spacing=spacing, projection=projection, dims=["latitude", "longitude"]
)
grid = vd.distance_mask(
(data.longitude, data.latitude),
maxdist=2 * spacing * 111e3,
grid=grid_full,
projection=projection,
)
# Calculate residuals between the predictions and the original input data. Even though
# we aren't using regularization or regularly distributed forces, the prediction won't
# be perfect because of the BlockReduce operation. We fit the gridder on the reduced
# observations, not the original data.
predicted = chain.predict(projection(*coordinates))
residuals = (data.velocity_east - predicted[0], data.velocity_north - predicted[1])
# Make maps of the original velocities, the gridded velocities, and the residuals
fig, axes = plt.subplots(
1, 2, figsize=(12, 8), subplot_kw=dict(projection=ccrs.Mercator())
)
crs = ccrs.PlateCarree()
# Plot the observed data and the residuals
ax = axes[0]
tmp = ax.quiver(
data.longitude.values,
data.latitude.values,
data.velocity_east.values,
data.velocity_north.values,
scale=0.3,
transform=crs,
width=0.001,
label="Velocities",
)
ax.quiverkey(tmp, 0.13, 0.18, 0.05, label="0.05 m/yr", coordinates="figure")
ax.quiver(
data.longitude.values,
data.latitude.values,
residuals[0].values,
residuals[1].values,
scale=0.3,
transform=crs,
color="r",
width=0.001,
label="Residuals",
)
ax.set_title("GPS horizontal velocities")
ax.legend(loc="lower left")
vd.datasets.setup_california_gps_map(ax)
# Plot the gridded data and the residuals
ax = axes[1]
tmp = ax.quiver(
grid.longitude.values,
grid.latitude.values,
grid.east_component.values,
grid.north_component.values,
scale=0.3,
transform=crs,
width=0.002,
)
ax.quiverkey(tmp, 0.63, 0.18, 0.05, label="0.05 m/yr", coordinates="figure")
ax.quiver(
data.longitude.values,
data.latitude.values,
residuals[0].values,
residuals[1].values,
scale=0.3,
transform=crs,
color="r",
width=0.001,
)
ax.set_title("Gridded velocities")
vd.datasets.setup_california_gps_map(ax)
plt.tight_layout()
plt.show()
Total running time of the script: ( 0 minutes 0.989 seconds)